Optimization essay
Almost every primer set which gives a good PCR product with a home-brew Taq enzyme can be optimized to purposes of HRMA.
Basic and usually sufficient procedure includes only testing different annealing temperatures in a range of 55-68°C with gradient PCR (you may try 50-60 and 60-70 as well if 55-68 does not work). After PCR it is important to check amplification with both LightScanner® and gel. The first shows how specific PCR was and the second allows to check both the specificity (ethidium bromide gel is less sensitive than HRMA) and the product size. The optimal assay does not give unspecific amplification and has acceptable amount of PCR product. Usually one row in the gradient cycler (12 wells) is enough to optimize assay for one primer set. On the picture below you can see results of an optimization procedure. The optimal Ta is 70°C.
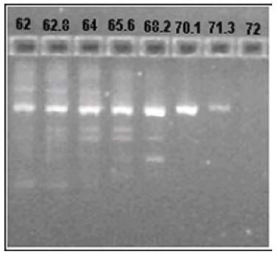
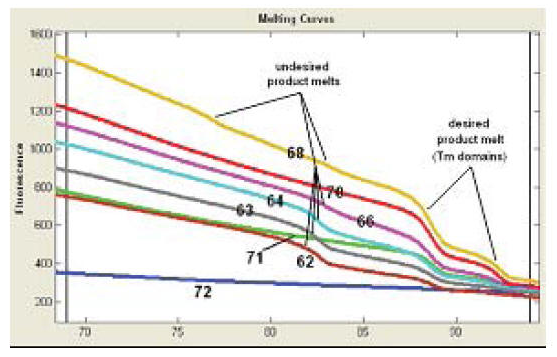
Some primer sets give lower amplification rates than others even after optimization. You can design new primers (preferred). You can also run another PCR (up to 40 cycles) with the same plate. You will notice that specific amplification will be detected after this second run. Afterwards you should put all ‘bad markers’ on one plate and ‘good markers’ on another plate. Then you can run long PCR for bad markers and normal (40-45 cycles) for good markers. Please check articles (1,2,3) for other solutions.
After the optimization fill the optimized settings in the file ‘list of optimized assays.xls’ on the LightScanner® PC (folder My documents/LightScanner documents/assays/list of optimized assays.xls). The new file is uploaded on the web site every week.
For the detailed description of the optimization check manual. It is also available in the software for data analysis and on the shelf behind the LightScanner®.